
PARIWISE is a transnational research project with partners from five different countries: Norway, Spain, Sweden, Tunisia and Uganda. The aim of the project is to study the role of wastewater treatment plants (WWTPs) in dispersal of antimicrobial resistance and how it influences grazing cattle and aquatic birds in contact with surface waters. It is a qualitative study on the abundance of antimicrobial resistant bacteria, resistance genes and antibiotic residues in waterbodies, grazing cattle and aquatic birds. In addition, the role of birds in transmission of antimicrobial resistant bacteria will be studied.
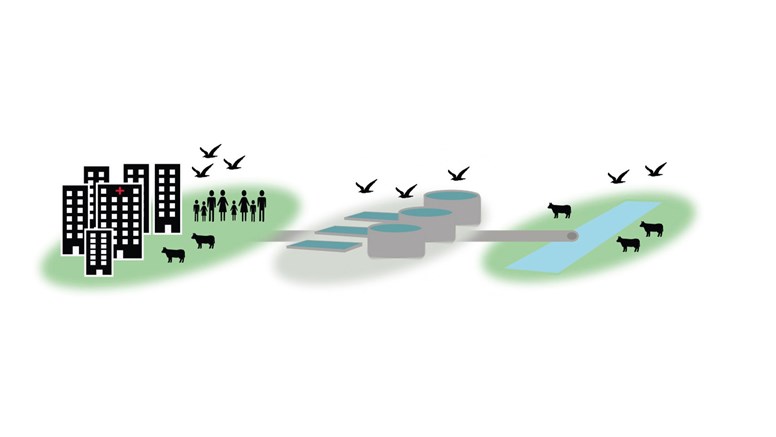
WWTPs play an important role in management of wastewater prior to dispersal into the environment. The environmental impact is dependent on local antibiotic use, environmental factors, and WWTP size5,6. It has also been shown that the presence of antimicrobial resistance genes decreases with increased distance to a WWTP7. The environment provides an immensely diverse microbiome that could potentially be acquired by bacteria with the ability to cause disease in humans and animals4.
The project truly embraces a One-Health approach by including aquatic environments linked to both human and animals. Grazing cattle and aquatic birds in close proximity to WWTPs could potentially acquire resistant bacteria from the surface water. Mutations usually arise during treatment as the selection pressure promotes difficult to treat bacteria; such a strong selection pressure is rarely seen in the environment4,8. On the other hand, agricultural runoffs and leaching from farms could also contaminate surface waters. Further, wild gulls have been proposed to transmit antimicrobial resistant bacteria due to their presence in human settings9,10.
References
-
- Global antimicrobial resistance and use surveillance system (GLASS) report 2021. Geneva: World Health Organization; 2021. Licence: CC BY-NC-SA 3.0 IGO.
- WHO Regional Office for Europe/European Centre for Disease Prevention and Control. Antimicrobial resistance surveillance in Europe 2022 – 2020 data.
- Swedres-Svarm 2020. Sales of antibiotics and occurrence of resistance in Sweden. Solna/Uppsala ISSN1650-6332.
- Larsson, D. G. J. & Flach, C.-F. Antibiotic resistance in the environment. Nat. Rev. Microbiol. 20, 257–269 (2022).
- Pärnänen, K. M. M. et al. Antibiotic resistance in European wastewater treatment plants mirrors the pattern of clinical antibiotic resistance prevalence. Sci. Adv. 5, eaau9124 (2019).
- Azuma, T. et al. Environmental fate of pharmaceutical compounds and antimicrobial-resistant bacteria in hospital effluents, and contributions to pollutant loads in the surface waters in Japan. Sci. Total Environ. 657, 476–484 (2019).
- McConnell, M. M. et al. Sources of Antibiotic Resistance Genes in a Rural River System. J. Environ. Qual. 47, 997–1005 (2018).
- Flach, C.-F., Genheden, M., Fick, J. & Joakim Larsson, D. G. A Comprehensive Screening of Escherichia coli Isolates from Scandinavia’s Largest Sewage Treatment Plant Indicates No Selection for Antibiotic Resistance. Environ. Sci. Technol. 52, 11419–11428 (2018).
- Atterby, C. et al. ESBL-producing Escherichia coli in Swedish gulls-A case of environmental pollution from humans? PloS One 12, e0190380 (2017).
- Ramey, A. M. et al. Antibiotic-Resistant Escherichia coli in Migratory Birds Inhabiting Remote Alaska. EcoHealth 15, 72–81 (2018).